|
CoV2ID provides a complete, quality checked and regularly updated list of oligonucleotides for SARS-CoV-2. The database evaluates available therapeutic and detection protocols according to the virus genetic diversity.
What can be done in CoV2ID
|
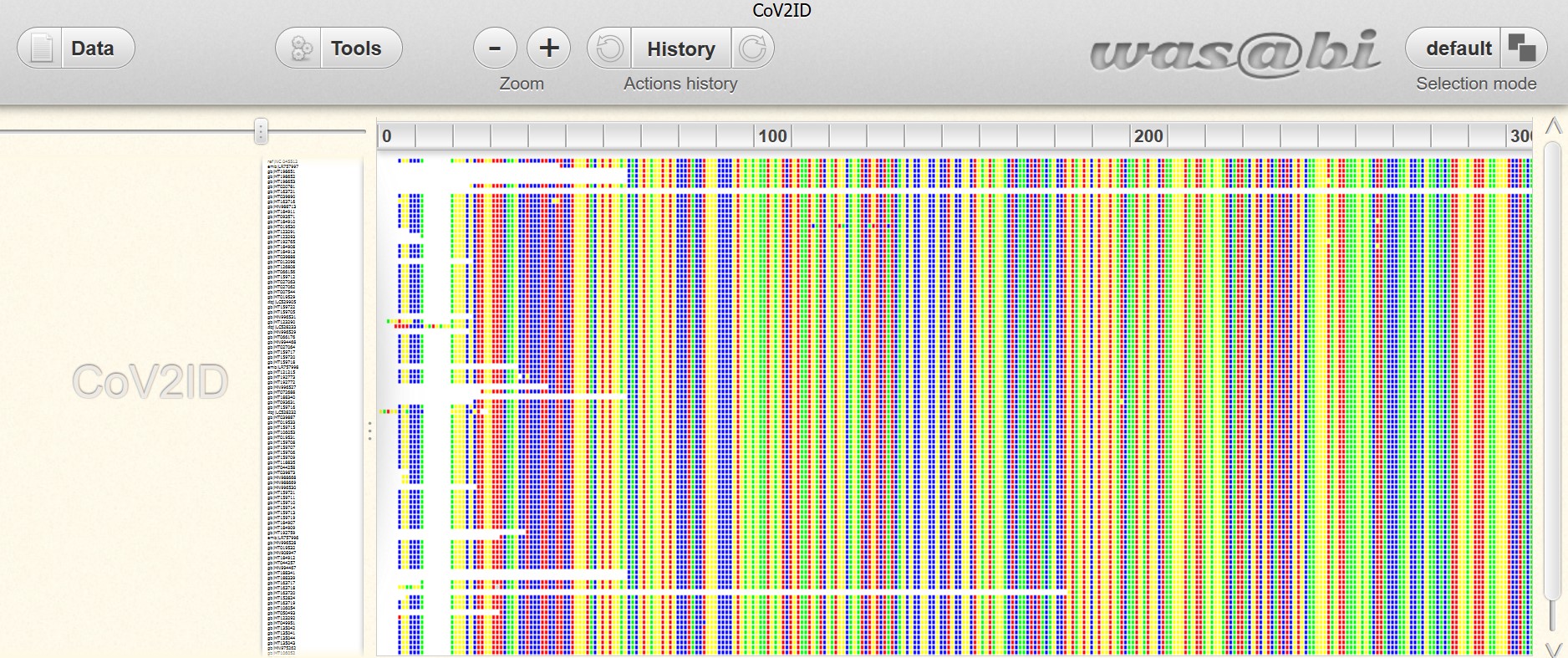
CoV2ID>> Find a brief description of the SARS-CoV-2 virus
|
|
Database content
| Total number of oligonucleotides: 492 | PCR primers: 181 | LAMP primers: 237 | Target Generation: 14 | Probes: 29 | Detection protocols: 57 |
 |
Localization of oligonucleotides (arrows) in the SARS-CoV-2 virus reference genome
|
What can be found in CoV2ID
Search for the best oligonucleotides according
to the degree of conservation
in specific sequence alignments
to the degree of conservation
in specific sequence alignments
Select the most conserved/variable regions
across the SARS-CoV-2 genome
across the SARS-CoV-2 genome
Find PCR primer pairs ordered by the average
degree of conservation
in diverse sequence alignments
degree of conservation
in diverse sequence alignments
Search for oligonucleotides described
in peer reviewed publications ordered
by the average degree of conservation
in diverse sequence alignments
in peer reviewed publications ordered
by the average degree of conservation
in diverse sequence alignments
Find information on the sequence conservation
measures used in this site
measures used in this site
Conserved
genomic regions
genomic regions
Select the best alignment according to
different measure of sequence conservation
different measure of sequence conservation
PCR primer pairs
The best oligonucleotides
Our sequence
conservation measures
conservation measures
The top alignments
Oligonucleotides
in specific alignments
in specific alignments
 |
Most conserved oligonucleotides for the SARS-CoV-2 genome
The CoV2ID score measures the level of conservation of the oligo according to the viral genetic diversity. The higher the score, the more conserved is the oligo.
| Top oligonucleotides | ||
| Databasereference | CoV2ID score | |
|---|---|---|
| CoV2ID001 | 100 | |
| CoV2ID006 | 100 | |
| CoV2ID008 | 100 | |
| CoV2ID047 | 100 | |
| CoV2ID050 | 100 | |
| CoV2ID054 | 100 | |
| CoV2ID073 | 100 | |
| CoV2ID075 | 100 | |
| CoV2ID086 | 100 | |
| CoV2ID158 | 100 | |
| CoV2ID160 | 100 | |
| CoV2ID161 | 100 | |
| CoV2ID165 | 100 | |
| CoV2ID167 | 100 | |
| CoV2ID174 | 100 | |
| Top PCR primers | ||
| Primer1 | Primer2 | CoV2ID score |
|---|---|---|
| CoV2ID008 | CoV2ID485 | 100 |
| CoV2ID174 | CoV2ID267 | 100 |
| CoV2ID178 | CoV2ID267 | 100 |
| CoV2ID193 | CoV2ID391 | 100 |
| CoV2ID267 | CoV2ID430 | 100 |
| CoV2ID358 | CoV2ID425 | 100 |
| CoV2ID410 | CoV2ID485 | 100 |
| CoV2ID459 | CoV2ID485 | 100 |
| CoV2ID485 | CoV2ID486 | 100 |
| CoV2ID324 | CoV2ID436 | 99.7 |
| CoV2ID054 | CoV2ID423 | 99.69 |
| CoV2ID160 | CoV2ID384 | 99.68 |
| CoV2ID178 | CoV2ID207 | 99.67 |
| CoV2ID001 | CoV2ID085 | 99.65 |
| CoV2ID050 | CoV2ID085 | 99.65 |
SARS-CoV-2 oligos in other human coronaviruses
Top divergent oligos in other coronaviruses(the best ones to avoid false positives) |
||
| Oligos | CoV2ID score | |
| CoV2ID362 | 8.43 | |
| CoV2ID195 | 8.49 | |
| CoV2ID232 | 8.86 | |
| CoV2ID479 | 8.95 | |
| CoV2ID193 | 9.02 | |
| CoV2ID249 | 9.08 | |
| CoV2ID191 | 9.13 | |
| CoV2ID469 | 9.22 | |
| CoV2ID199 | 9.25 | |
| CoV2ID320 | 9.31 | |
Top conserved oligos in other coronaviruses(the most similar, may cause false positives) |
||
| Oligonucleotide | CoV2ID score | |
| CoV2ID102 | 91.43 | |
| CoV2ID280 | 89.33 | |
| CoV2ID331 | 87.54 | |
| CoV2ID330 | 83.6 | |
| CoV2ID272 | 83.08 | |
| CoV2ID333 | 82.4 | |
| CoV2ID332 | 81.74 | |
| CoV2ID455 | 81.12 | |
| CoV2ID058 | 79.05 | |
| CoV2ID105 | 78.1 | |
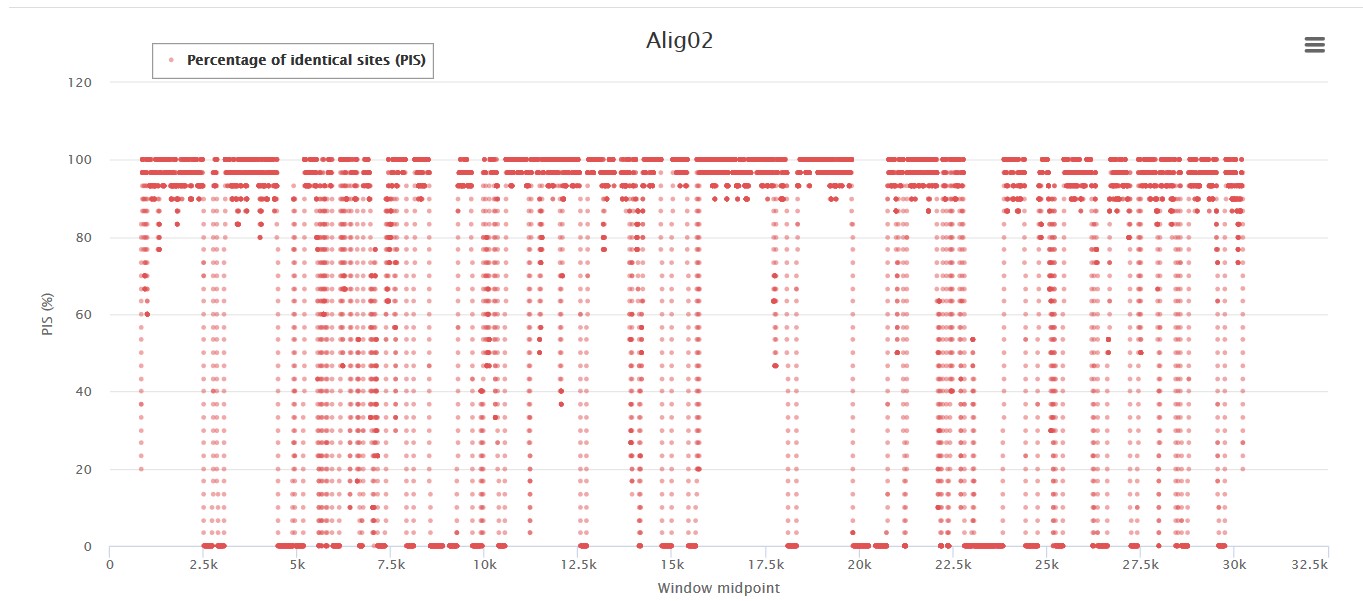
Most conserved regions for the SARS-CoV-2 genome
The percentage of identical sites and pairwise identity level of conservation of the oligo according to genetic diversity. The higher the score, the more conserved is the oligo.
List of the most conserved genomic regions considering the PIS and PPI (100 nt windows):
| Top region (PIS) | ||
| Genome Position | PIS | |
|---|---|---|
| 15901-16000 | 100 | |
| 15951-16050 | 100 | |
| 5551-5650 | 99 | |
| 8651-8750 | 99 | |
| 15301-15400 | 99 | |
| 24551-24650 | 99 | |
| 26351-26450 | 99 | |
| 8701-8800 | 98 | |
| 9951-10050 | 98 | |
| 15201-15300 | 98 | |
| 15251-15350 | 98 | |
| 19601-19700 | 98 | |
| 26251-26350 | 98 | |
| 26301-26400 | 98 | |
| 26401-26500 | 98 | |
| Top region (PPI) | ||
| Genome Position | PPI | |
|---|---|---|
| 1101-1200 | 100 | |
| 2501-2600 | 100 | |
| 2901-3000 | 100 | |
| 2951-3050 | 100 | |
| 3401-3500 | 100 | |
| 3701-3800 | 100 | |
| 3751-3850 | 100 | |
| 3801-3900 | 100 | |
| 3851-3950 | 100 | |
| 4751-4850 | 100 | |
| 5051-5150 | 100 | |
| 5101-5200 | 100 | |
| 5151-5250 | 100 | |
| 5301-5400 | 100 | |
| 5551-5650 | 100 | |